(Note: cette page n'existe qu'en anglais - Nota: deze pagina bestaat enkel in het Engels)
The deployment of oligonucleotides and engineered nucleases (Meganucleases, Zinc finger nucleases, TALENs and the CRISPR/cas system) enabled a significant breakthrough in targeted genome editing and offers an unprecedented way of modifying the genome of organisms in an efficient and cost-effective way.
The SBB has been following the developments and applications in this field (Breyer et al., 2009; Pauwels et al., 2014, van der Vlugt, et al., 2018) and has been involved in several initiatives at the European and Belgian level in the past years related to the clarification of the legal status of the organisms obtained by targeted genome editing and to the assessment of the possible challenges in terms of risk assessment, detection and monitoring. In 2016, the SBB issued scientific considerations on the use of CRISPR/cas system upon request of the competent authorities. With the ruling of the European Court of Justice in July 2018, the legal status of organisms obtained by targeted genome editing has been partially clarified at European level (see regulatory considerations). However, the SBB considers that the scientific considerations it expressed in 2016 remain valid.
Are you using engineered nucleases in the context of your activities? The following aspects could be relevant to perform a risk assessment:
- The predictability and safety of the approach.
- The characteristics of several nucleases and systems
- The molecular mechanisms (mode of action) and the different possible outcomes, commonly categorized as SDN-1, SDN-2 and SDN-3.
- The different modes of delivery of the engineered nucleases.
Find out more on how CRISPR platforms can be used to generate gene drives or to regulate gene expression.
All the references mentioned are gathered here.
Engineered nucleases: Characteristics
Meganucleases are based on naturally occuring endodeoxyribonucleases recognizing long (>12 bp) DNA sequences. Contrary to other nucleases such as Zinc-finger nucleases or TALENs, Meganucleases present no modular structure, therefore the redesign of DNA-recognition moieties is relatively fastidious. Nevertheless, some research groups and biotech companies managed to engineer these naturally occurring proteins in order to direct them to a locus of interest.
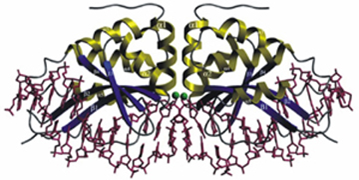
I-CreI endonuclease. Adapted from Grizot et al., NAR, 2009, 37(16)
Zinc-Finger Nucleases are hybrid proteins with a DNA-recognition domain made up of zinc fingers (ZFs) and a DNA-cleavage domain coming from the FokI restriction endonuclease. Each ZF interacts mainly with 3 base pairs (bp) of DNA. The DNA-binding specificities of each ZF can easily be altered. Engineered ZFs have been combined to recognize 9-, 12-, 15- or 18-bp DNA sequences.
The FokI domain must dimerize to obtain an active nuclease, which means that the design of ZFNs includes the design of a pair of ZFN monomers.
To circumvent off-target activity and ZF toxicity upon homodimerization of the ZFN monomers, the design of the nucleases consists in altering the dimeric interface so as to avoid the formation of homodimeric forms and to promote the heterodimeric forms. Incompatibilities at the finger-finger interface (known as context-dependent effects) may also occur and lead to decreased or altered DNA-binding specificity. Design approaches explicitly account for these context-dependent effects between adjacent fingers.
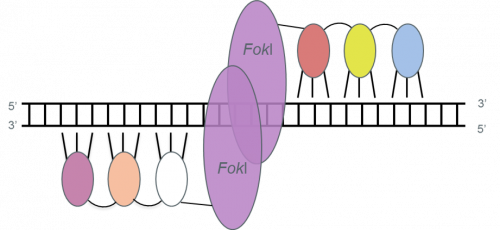
ZFNs. Each zinc finger (small ovals) in a zinc-finger nuclease (ZFN) binds primarily to three consecutive base pairs. Different colors indicate fingers recognizing different DNA triplets. Each set of fingers is joined to a FokI-derived cleavage domain (large ovals) by a short linker. The target site is composed of two target halfsites. ZFN pairs can be designed to bind genomic sequences of 18–36 nucleotides long. Adapted from Pauwels et al., New Biotechnology (2014), 31(1), 18-27.
TALENs have a modular structure with the Fok I restriction endonuclease as DNA-cleavage domain. Unlike ZFNs the DNA-binding domain of TALENs is composed of a variable number of 34/35 amino acid repeats, in which a ‘repeat-variable di-residue’ (RVD) determines the binding with a single base pair of DNA. There is a one-to-one recognition between the RVD module and the single base pair, which simplifies the engineering of specific DNA-binding domains.
One limitation seems to be the requirement of having a thymine at the 5’ end of the target sequence. The construction of the tandem arrays can also be challenging because TALENs could consist of up to 20 RVD and these highly homologous sequences can recombine with each other in cells.
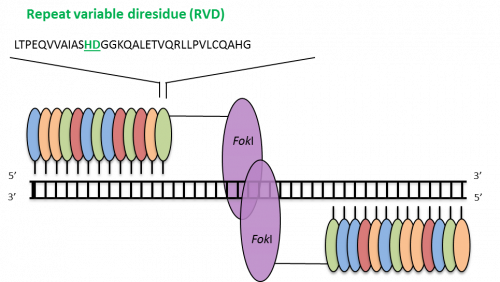
TALENs. Each module (small ovals) binds a single base pair; the four colors indicate modules for each of the four base pairs. The minimum effective number of modules per monomer is 10–12, but more are typically used. Adapted from Pauwels et al., New Biotechnology (2014), 31(1), 18-27.
The CRISPR/Cas system is the most recent platform developed in the field of genome editing. CRISPR stands for ‘Clustered Regularly Interspersed Short Palindromic Repeats’ and Cas for the Cas endonuclease. Unlike all other platforms for engineered nucleases, the DNA-recognition is based on RNA-DNA interactions. This enables a faster and more cost-effective engineering of the DNA-recognition moiety. Other critical elements are the CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracr RNA), which are usually expressed as a single RNA (guide RNA – gRNA). The gRNA binds Cas9 nuclease and directs cleavage to a target sequence in DNA that has homology to the RNA 5’ end. The cleavage by Cas9 is highly dependent on the presence of a two base pair sequence adjacent to the region of homology, called protospacer adjacent motif (PAM), which is recognized by the complex. A particularity of this nuclease system is the capacity to perform multiplexed targeting (simultaneous editing of several sites) by encoding multiple guide sequences into a single CRISPR array.
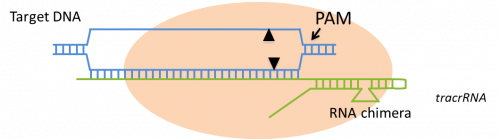
The CRISPR minimal-cleavage elements. A single RNA chimera (green lines) with the critical elements of the crRNA and tracrRNA binds Cas9 protein (orange oval) and directs cleavage (arrowheads) to a sequence in DNA (blue) that has homology to the RNA 5’ end. The region of RNA–DNA base pairing provides cleavage specificity. The target must have a particular two-base-pair sequence adjacent to the region of homology, called PAM, which is recognized by the complex. Adapted from Pauwels et al., New Biotechnology (2014), 31(1), 18-27
Alternative CRISPR system are being investigated because of their smaller size and different sequence requirements compared to Cas9 (e.g. Cpf1 (Kim et al., 2016; Kleinstiver et al., 2016) for targeting AT-rich genomes) or because the enzyme targets RNA rather than DNA (e.g. C2c2 (Abudayyeh et al., 2016)). These developments illustrate how the CRISPR system allows for some flexibility that can be harnessed for various gene editing technologies.
Engineered nucleases: Molecular mechanisms and possible outcomes
The mode of action of so-called Site Directed Nucleases (SDN) such as Meganucleases, Zinc-Finger Nucleases, TALENs and the CRISPR-Cas system is always based on the site-specific cleavage of the DNA by means of a nuclease and the triggering of the cell’s gene repair mechanisms of the cell, through non-homologous end-joining (NHEJ) or homologous recombination (HR). These repair mechanism result in the intended genetic modifications ranging from a targeted point mutation to large deletions or insertions of exogenous DNA at predefined sites in the genome.
The outcome of genome editing using engineered nucleases in terms of efficiency and accuracy depends on various factors such as the choice of the type of engineered nuclease, the method of delivery, the transient/stable expression of SDN compounds (quantity), the type of DNA breakage (double-strand breaks (DSB), nicks or double nicks), the number of DSB introduced (whether or not the DSB is unique), and the species-dependent differences in preponderance of NHEJ compared to HDR (e.g. for CRISPR/cas system reviewed in Bortesi et al., 2016).
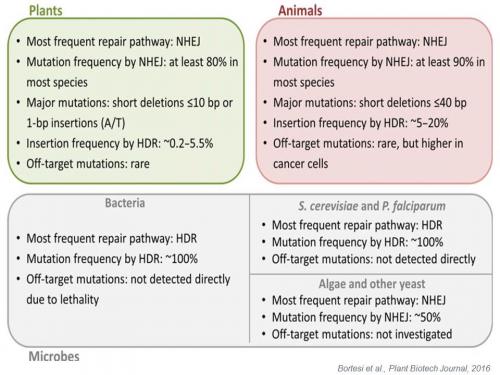
The outcome of genome editing is subject to species-dependent effects determined by the prevalent DNA repair pathways. Adapted from Bortesi et al., Plant Biotechnology Journal (2016), 14, 2203-2216.
Within the framework of ongoing discussion on the legal status of organisms obtained by the use of SDN, applications are generally grouped in three categories depending on the cell’s gene repair mechanism triggered and the delivery or not of a stretch of exogenous donor DNA.
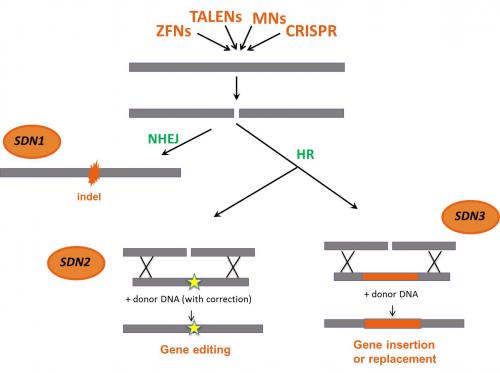
- In SDN-1 applications, mutations consisting of changes in a few base pairs, short deletions or insertions (indels) are generated in a predefined region in the genome as a result of an error-prone gene repair mechanisms of the cell (NHEJ). The repair mechanism does not require exogeneous delivered DNA.
- In SDN-2 applications, specific point mutations, small deletions/additions are generated as a result of the introduction into the cell of a repair DNA template (donor DNA) homologous to the targeted area. By means of homologous recombination (HR), precise and small genetic modification can be achieved.
- In SDN-3 applications, entire genes can be inserted into a desired site in the genome. This is enabled by the delivery of a large stretch of recombinant DNA molecule (an exogenous donor DNA up to several kilobases long). The insertion can take place either by HR or by NHEJ.
At the molecular level, the result of genetic modifications obtained by SDN-1 and SDN-2 are similar to what can be obtained by chemical mutagenesis, by irradiation or by spontaneous natural mutations, and are not distinguishable from them. The genetic modifications are a result of the cellular DNA repair mechanisms of the host that also repair DNA breaks as a result of naturally occurring processes.
Engineered nucleases: Mode of delivery
Nucleases can be brought into cultured cells, one-cell embryos or whole organisms in various forms, including
- plasmid DNA, (electroporation or liposome transfection of genes encoding engineered nucleases)
- in vitro transcribed mRNA (micro-injection into one-cell embryos to create knockout organisms). Compared with plasmid DNA transfection, mRNA delivery of programmable nucleases can lead to faster expression and avoid unwanted integration into the cell genome of plasmid DNA that encodes the nucleases.
- Non-integrating viral vectors — such as integrase-deficient lentivirus vectors (IDLVs), adenoviruses and adeno-associated viruses (AAVs) — can deliver genes encoding programmable nucleases both in vitro and in vivo. Sequences of ZFNs are the most compact for being packaged into small vectors. Unfortunately, the construction of IDLVs with TALENs can be difficult because highly homologous TALE repeats often lead to unwanted recombination in cells.
- Integrating vectors such as lentiviral vectors have been used for continuous expression of Cas9 and sgRNAs in mammalian cells, and have resulted in almost 100% mutation rates at target sites. However, this method is likely to increase off-target effects.
Safe and efficient delivery of nucleases into target cells is one of the main obstacles towards their use in the area of gene therapy. Later advances focus on non-viral delivery of genome-editing nucleases such as the delivery of Cas9 proteins complexed with sgRNA, so-called Cas9/sgRNA ribonucleoproteins (see this review). Unlike DNA-based expression, protein delivery avoids unwanted incorporation of foreign DNA. It may also reduce off-target effects because, unlike plasmid DNA, proteins are rapidly degraded in cells.
Targeted genome editing: Safety and risk assessment considerations
Before the era of targeted genome editing, the introduction of novel traits in organisms by genetic engineering mostly relied on the use of techniques of stable yet random introduction of the genetic modification (e.g. conventional mutagenesis) or foreign genetic material (transgenesis). Due to this randomness, unwanted alterations in the genome such as (unwanted) disruption of genes and/or regulatory elements or the creation of new undesirable open reading frames (e.g. with a sequence similar to known toxins or allergens) can occur explaining why screening and selection processes of GMOs with desirable traits are often cumbersome and time-consuming. The use of site-directed nucleases (SDN) offers the possibility to reduce the probability of these unwanted adverse effects, as it provides a way to target a predefined locus of the genome. In the field of plant and animal breeding, this approach provides an unprecedented way of introducing agronomical interesting traits in a more efficient and cost-effective way compared to traditional breeding.
Within this perspective, genome editing is a more targeted and predictable approach than classical mutagenesis, where modifications are scattered all over the genome, or than Agrobacterium tumefaciens-mediated transformation, where gene cassettes are inserted in a random way in the genome of a plant or tissue culture that cause somatic mutations (Li et al., 2016). Notably, these arguments brought EFSA to conclude that less event-specific data may be required for plants developed by the SDN-3 approach, (which is an approach where foreign DNA is targeted by means of the action of SDN and inserted by means of homologous recombination).
| Off-target activity Numerous experiments were conducted to study off-target mutations associated to the use of SDN in human cell, animals, plants and microbes (e.g. reviewed in Bortesi et al., 2016) and several approaches have been described to increase targeting accuracy of SDN by decreasing the likelihood of off-target cleavage, including for instance:
|
Where SDN is used to perform small deletions or point mutations (SDN-1 or SDN-2 approach), it is currently a matter of debate to what extent off-target events need to be detected and characterized. In this context the impact of an off-target event should be considered in light of the organism and its application.
In plants, it is important to consider the context of natural causes of genetic variability and the frequency of indels occurring spontaneously or after chemical mutagenesis (Ladics et al., 2015). Differences in genetic background may have an impact on whether an induced DNA change may have an effect on the characteristics of the product/organism. There are also naturally occurring events that lead to what is called plant genome plasticity. For example in maize it is known that transposons (transposable elements) can move their location into the genome. Transposon insertions may have an impact on gene expression and on how the maize plants interact with their environment (e.g. drought tolerance, flowering time, ability to grow on soil with altered composition).
Along with the natural causes of genetic variability, it is not an easy task to set-up a reference genome to which an organism modified by genome editing should be compared to, in order to assess the effect of an off-target event. Notably, with SDN-induced small deletions or mutations, it is technically not possible to distinguish off-target events from naturally occurring changes in the genome.
Last but not least it is a common practice in plants to have off-target events resulting in unintended mutations be segregated away during the selection and breeding process.
In animal breeding, where selection steps are more laborious, targeted genome modification would be beneficial provided that the effect of off-target events are assessed more closely (note that the EFSA guidance on the environmental risk assessment of GM animals includes aspects of animal health and welfare). For gene therapy applications, risk assessment at large takes into account patient safety and risk/benefit balance considerations. In the latter case, an assessment of off-target events may outweigh pure biosafety aspects considered within the framework of GM legislation.
Regulatory considerations
Genome editing techniques involving the use of Site-Directed-Nucleases (SDN) make it possible to induce modifications in a pre-defined region of the genome without the need to introduce foreign (exogenous) DNA. In most of the cases the resulting organisms cannot be distinguished from those generated through traditional breeding or classic mutagenesis. Notably, organisms developed through classical mutagenesis fall outside the scope of the current EU regulatory system for GMO. Some applications of SDN-mediated targeted genome editing and ODM are therefore blurring the boundaries of the current GM regulation in the European Union.
A New Plant Breeding Techniques working group set up by the European Commission (EC) in 2007 was mandated to evaluate eight plant breeding techniques including oligo-directed mutagenesis (ODM) and SDN-mediated editing. While addressing how these techniques relate to the European GMO legislation, a majority of experts of this working group was of the opinion that ODM and SDN-1 and SDN-2 are a mutagenesis technique that could be exempted from the EU GMO regulation on the condition that the nuclease is not stably expressed from a recombinant nucleic acid molecule.
Meanwhile a number of authorities or advisory bodies of EU member states (ACRE, 2013; BVL, 2017; HCB, 2017; COGEM, 2017; SWE, 2015) have concluded that organisms obtained by ODM, SDN-1 and SDN-2 approaches should be considered for exclusion from the GMO legislation. For others a precautionary approach should be adopted, taking into account the uncertainties and limited knowledge on the mode of action of certain types of modifications (Eckerstorfer et al., 2014; opinion commissioned by the German Federal Agency for Nature Conservation, Case C-528/16 Confédération Paysanne and Others).
|
|
Numerous official institutions, advisory bodies and scientific authorities (e.g. UK House of Commons Science and Technology Committee, Germany’s National Academy of sciences, European Academies’ Science Advisory Council, European Plant Science Organisation) voiced that GMO regulation in the EU should be revisited to be more clearly product-oriented, so that an organism should be regulated and the risk should be assessed on the basis of its phenotype (e.g. specific agricultural trait) instead of the technique used to develop the organism (process-oriented). Several scientists also issued similar opinions in this regard (e.g. Carroll et al., 2016; Pacher and Puchta, 2017; Ricroch et al., 2016).
The European Commission has postponed several times a decision on the legal status (in or out of scope of the EU GMO legislation) of organisms developed by new breeding techniques, including ODM and SDN-mediated editing methods. In October 2016 a French court case of anti-GMO groups against the French government was brought to the European Court of Justice (ECJ), containing the request for interpretation of EU law by determining whether organisms obtained by mutagenesis are GMO’s and fall within the scope of Directive 2001/18 (Case C-528/16 Confédération Paysanne and Others). In January 2018, an Advocate General of the Court of Justice of the EU (ECJ) published his legal opinion clarifying how mutagenesis techniques, including directed mutagenesis and applications of gene editing, should be regulated under EU law . It concludes (amongst others) that ‘The exemption laid down in Article 3(1) of Directive 2001/18, read in conjunction with its Annex I B covers all organisms obtained by any technique of mutagenesis, irrespective of their use at the date of the adoption of that directive, on the condition that they do not involve the use of recombinant nucleic acid molecules or genetically modified organisms other than those produced by one or more of the methods listed in Annex I B‘. An Advocate General’s legal opinion is non-binding and advisory, and was indeed not followed by the Court. The judgement, issued on 25 July 2018, ruled that mutagenesis techniques developed since the adoption of the Directive 2001/18 are subject to this Directive, as they cannot be considered as having a long record of safe use. It also affirms the process-oriented interpretation of the Directive.
In the USA, genetically engineered plants are regulated by the USDA, and by the EPA when they may have pesticidal properties. Unlike transgenic plants modified using conventional techniques, gene edited plants involving the use of CRISPR-Cas9 do not require USDA oversight because the resulting plants don’t contain DNA from “plant pests” such as viruses or bacteria. Other plant applications using other gene-editing techniques, such as TALEN and ZFN transcription, can also fall outside of USDA’s authority (Wolt et al., 2016).
GM animals are in large part regulated by the FDA. In 2017, the FDA made available draft rules stating that animals with intentionally altered genomes would be subject to safety testing similarly to new drugs (DHHS–FDA 2017). In the same period however, the USDA released draft rules that would include genome editing as a type of genetic engineering, but exclude SDN-1 edits from regulation as “genetically engineered” (USDA–APHIS 2017a).
Engineered nucleases: Applications
The use of nucleases directed to cleave the genome at specific sites of interest was first applied in Drosophila (Bibikova et al.,2002). Until 2014, the three classes of nucleases most used were ZFNs, TALENs and meganucleases (also called homing endonucleases). These three classes have been largely superseded by the CRISPR system since its application in genome editing. The number of scientific papers published in 2015 related to CRISPR almost tripled compared with combined number of publication output for the three other classes of nucleases.
There are numerous examples where gene editing approaches are used to better understand gene function in genomes of human somatic cells, viruses, bacteria, animals and plants. The deployment of gene editing tools has also been described in the field of plant biotechnology, animal breeding and human health applications.
It is impossible to provide an exhaustive and updated list of applications. The examples below therefore only give an indication of some interesting outcomes:
- The genetic modification of economically important crops such as soybean with drought and salt tolerance, camelina with increased oil content, mushrooms and apples with anti-browning properties, low phytic acid corn, rice resistant against blight or wheat plants resistant against powdery mildew. Notably, novel traits useful for improving sustainability can be obtained by targeted deletions (SDN-1 approach) (reviewed by van de Wiel et al). SDN-1 induced mutations can also be used to improve the efficiency of biofuel production by decreasing the amount of lignin in biofuel feedstock crops (Jung and Altpeter, 2016).
- The genetic modification of animals, not only for creating animal models to study human disease (e.g. Ji et al., 2015) or to alter traits of pets (Reardon 2016) but also to improve production traits in livestock such as increased muscling for meat production in pigs (Rao et al., 2016), sheep and cattle (Proudfoot et al., 2015) or to produce hornless dairy cattle.
- Alternative sources for organs and tissues for pig-to-human xenotransplantation by the inactivation of porcine endogenous retroviruses in pigs.
- Alternative therapeutic strategies for infectious diseases such as efforts towards clearance of HIV reservoirs.
- De-extinction projects aiming at recreating extinct species like the passenger pigeon (Ectopistes migratorius) or the woolly mammoth based on editing of an Asian elephant (Elephas maximus) genome (ref).
- Vector control (gene drives).
- Human gene therapy as exemplified by three phase I clinical trials aiming at treating HIV/AIDS patients.
- Germline engineering as exemplified by three studies reporting the use of CRISPr-Cas9 in diploid human embryos.
- The development of microalgae as third generation biofuels feedstocks.
Gene drives
Along with the possibility to apply the CRISPR/Cas technology in virtually all possible organisms, Esvelt et al. described in 2014 the potential of RNA-guided gene drives based on CRISPR/Cas system to spread altered traits through wild populations in a non-Mendelian way. Indeed, the versatility of the CRISPR/Cas system opens a lot of new avenues in domains such as human health, agriculture and environment. Gantz and Bier provided the first proof of principle in Drosophila (figure below). They integrated the Cas9–sgRNA cassette into the genome (yellow bar) in such a way that it is flanked by the segments of the target sequences of the endonuclease (blue bar).
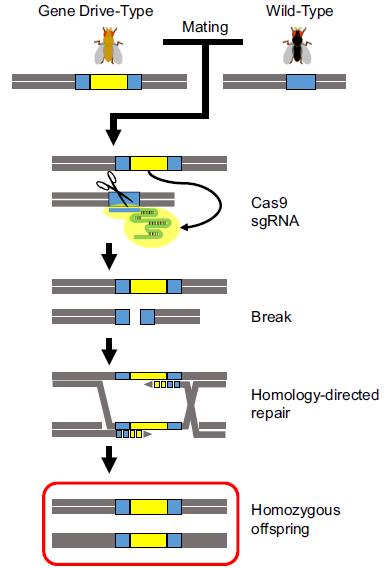
When an organism with a gene drive construct mates with a wild-type organism (sexual reproduction), the gene drive is inherited in a non-Mendelian way. Adapted from Schenkel and Leggewie. J.Verbr.Lebensm. (2015) 10: 263-268.
When the endonuclease is expressed, a double-strand break (DSB) takes place in the target sequence. In case homologous recombination is used, the CRISPR/Cas-encoding cassette is integrated at the cleavage site, resulting in a homozygous mutation that can be inherited in all offspring. An essential part of the ability to construct gene drives relies on the DNA-repair mechanism of the cell. If the DSB is repaired by non-homologous end-joining, another predominant DNA-repair mechanism present in various cell types, no copy of the CRISPR/Cas cassette can be inserted and no gene drive will be created.
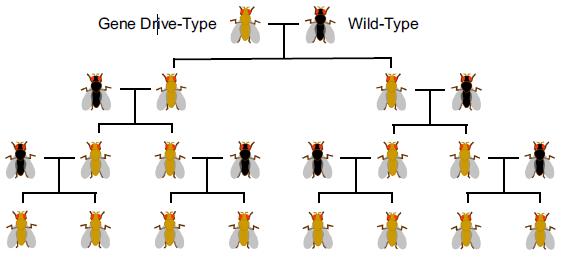
Numerous papers in scientific (and mainstream) journals have pointed towards the risk associated with the potential of gene drives to alter entire wild populations, as it could have an effect on the environment or ecosystems.
In order to conduct a proper risk assessment, it is essential to assess the effectiveness with which a gene drive could be introduced (and maintained) into a population. Following aspects may be considered:
- Presence of wild populations and probability of interaction;
- Generation time : A 100% effective gene drive can only ever double in frequency with each generation inheriting the drive mechanism. Mosquitoes have an average generation time of three weeks and it would take several generations to spread a gene drive to a portion of a local mosquito population. By comparison, a viral pandemic would affect national and international populations in a period of weeks.
- The cell’s predominant DNA repair mechanisms (only homologous recombination ensures that a copy of the CRISPR/Cas cassette is inserted);
- Fitness cost of the introduced trait;
- Development of resistance mechanisms: natural genetic variation may hamper the recognition of target sites by the endonuclease;
- The effectiveness of ‘built-in’ control strategies.
Control strategies may rely on containment measures (physical barriers) and/or on the adoption of ‘in build’ approaches by designing genetic constructs that allow to correct or reverse genomic alterations made by an earlier drive (Esvelt et al., 2014).
Gene drive organisms are GMOs and thus subject to the GMO regulatory framework and its provisions. Several agencies and/or organizations have issued recommendations on the responsible and safe use of gene drives, including RIVM, ZKBS, HCB, Australian academies of Science and National Academies of Science, Engineering and Medicine.
The SBB also co-authored the following paper: ‘A framework for the risk assessment and management of Gene Drive Technology in contained use’ (van der Vlugt et al., 2018).
Epigenetic regulation
TALEN and CRISPR platforms are not only being used as a tool for genome editing. The DNA-binding moieties of these platforms can also be fused with effectors such as activators, histone demethylases or acetylases instead of nucleases. This opens another area of possibilities in the field of gene regulation and epigenome engineering.
Nature video: CRISPR: gene editing and beyond
For example, the DNA binding moiety can be coupled to switches controlled by certain triggers such as chemicals or light for spatial, temporal and reversible control of gene regulation. These so-called optogenetics studies may be helpful to understand dynamic gene networks.
Another approach for gene regulation consists in the retargeting of the dsDNA binding capacity of Cas9 to (RNA guided) ssRNA binding/cleavage, thereby offering an alternative to RNA interference.
These developments illustrate how the CRISPR system allows for some flexibility that can be harnessed for various technologies (reviewed by Dominguez et al., 2016).
